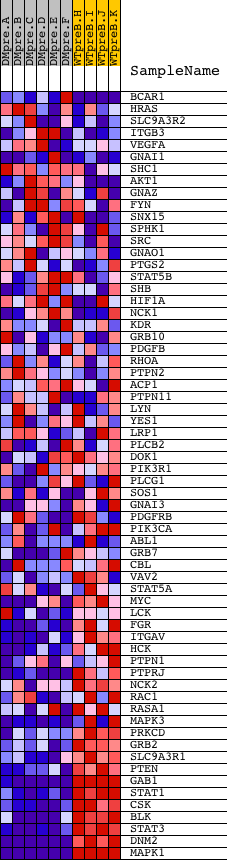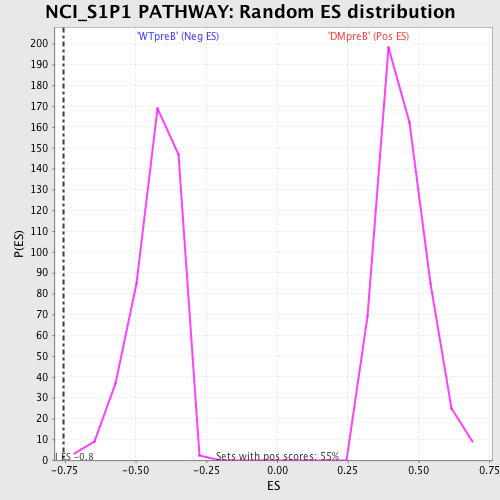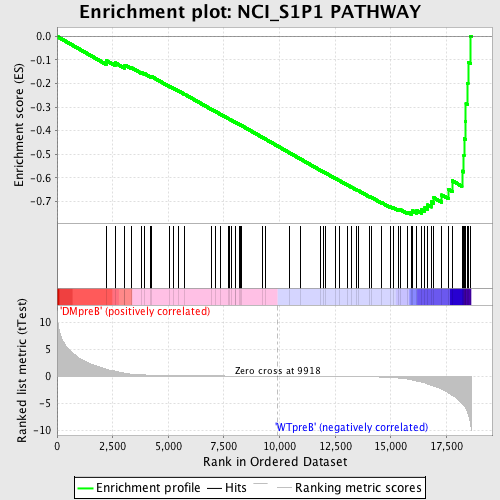
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NCI_S1P1 PATHWAY |
| Enrichment Score (ES) | -0.7561022 |
| Normalized Enrichment Score (NES) | -1.7613945 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0053009074 |
| FWER p-Value | 0.022 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BCAR1 | 18741 | 2225 | 1.277 | -0.1033 | No | ||
| 2 | HRAS | 4868 | 2610 | 0.906 | -0.1122 | No | ||
| 3 | SLC9A3R2 | 23089 1544 1514 | 3024 | 0.543 | -0.1274 | No | ||
| 4 | ITGB3 | 20631 | 3043 | 0.531 | -0.1215 | No | ||
| 5 | VEGFA | 22969 | 3350 | 0.369 | -0.1332 | No | ||
| 6 | GNAI1 | 9024 | 3780 | 0.255 | -0.1530 | No | ||
| 7 | SHC1 | 9813 9812 5430 | 3936 | 0.225 | -0.1584 | No | ||
| 8 | AKT1 | 8568 | 4218 | 0.183 | -0.1712 | No | ||
| 9 | GNAZ | 71 | 4228 | 0.182 | -0.1693 | No | ||
| 10 | FYN | 3375 3395 20052 | 5072 | 0.109 | -0.2134 | No | ||
| 11 | SNX15 | 23993 3761 | 5222 | 0.100 | -0.2201 | No | ||
| 12 | SPHK1 | 5484 1266 1402 | 5473 | 0.088 | -0.2324 | No | ||
| 13 | SRC | 5507 | 5727 | 0.076 | -0.2451 | No | ||
| 14 | GNAO1 | 4785 3829 | 6926 | 0.044 | -0.3091 | No | ||
| 15 | PTGS2 | 5317 9655 | 7102 | 0.040 | -0.3180 | No | ||
| 16 | STAT5B | 20222 | 7325 | 0.036 | -0.3295 | No | ||
| 17 | SHB | 10493 | 7714 | 0.029 | -0.3500 | No | ||
| 18 | HIF1A | 4850 | 7746 | 0.029 | -0.3513 | No | ||
| 19 | NCK1 | 9447 5152 | 7819 | 0.028 | -0.3548 | No | ||
| 20 | KDR | 16509 | 8027 | 0.024 | -0.3657 | No | ||
| 21 | GRB10 | 4799 | 8036 | 0.024 | -0.3658 | No | ||
| 22 | PDGFB | 22199 2311 | 8207 | 0.022 | -0.3747 | No | ||
| 23 | RHOA | 8624 4409 4410 | 8261 | 0.021 | -0.3773 | No | ||
| 24 | PTPN2 | 23414 | 8306 | 0.020 | -0.3794 | No | ||
| 25 | ACP1 | 4317 | 9230 | 0.009 | -0.4290 | No | ||
| 26 | PTPN11 | 5326 16391 9660 | 9360 | 0.007 | -0.4359 | No | ||
| 27 | LYN | 16281 | 10463 | -0.007 | -0.4952 | No | ||
| 28 | YES1 | 5930 | 10960 | -0.014 | -0.5217 | No | ||
| 29 | LRP1 | 9284 | 11848 | -0.027 | -0.5692 | No | ||
| 30 | PLCB2 | 5262 | 11963 | -0.029 | -0.5750 | No | ||
| 31 | DOK1 | 17104 1018 1177 | 12045 | -0.031 | -0.5789 | No | ||
| 32 | PIK3R1 | 3170 | 12502 | -0.041 | -0.6030 | No | ||
| 33 | PLCG1 | 14753 | 12675 | -0.046 | -0.6116 | No | ||
| 34 | SOS1 | 5476 | 13046 | -0.056 | -0.6308 | No | ||
| 35 | GNAI3 | 15198 | 13219 | -0.062 | -0.6393 | No | ||
| 36 | PDGFRB | 23539 | 13453 | -0.070 | -0.6510 | No | ||
| 37 | PIK3CA | 9562 | 13540 | -0.075 | -0.6546 | No | ||
| 38 | ABL1 | 2693 4301 2794 | 14050 | -0.106 | -0.6807 | No | ||
| 39 | GRB7 | 20673 | 14124 | -0.112 | -0.6832 | No | ||
| 40 | CBL | 19154 | 14602 | -0.166 | -0.7067 | No | ||
| 41 | VAV2 | 5848 2670 | 14978 | -0.236 | -0.7239 | No | ||
| 42 | STAT5A | 20664 | 15113 | -0.268 | -0.7276 | No | ||
| 43 | MYC | 22465 9435 | 15334 | -0.330 | -0.7352 | No | ||
| 44 | LCK | 15746 | 15440 | -0.368 | -0.7360 | No | ||
| 45 | FGR | 4723 | 15758 | -0.526 | -0.7463 | No | ||
| 46 | ITGAV | 4931 | 15941 | -0.673 | -0.7474 | Yes | ||
| 47 | HCK | 14787 | 15979 | -0.695 | -0.7403 | Yes | ||
| 48 | PTPN1 | 5325 | 16168 | -0.897 | -0.7388 | Yes | ||
| 49 | PTPRJ | 9664 | 16387 | -1.089 | -0.7364 | Yes | ||
| 50 | NCK2 | 9448 | 16530 | -1.250 | -0.7278 | Yes | ||
| 51 | RAC1 | 16302 | 16651 | -1.434 | -0.7156 | Yes | ||
| 52 | RASA1 | 10174 | 16815 | -1.689 | -0.7024 | Yes | ||
| 53 | MAPK3 | 6458 11170 | 16931 | -1.809 | -0.6851 | Yes | ||
| 54 | PRKCD | 21897 | 17290 | -2.395 | -0.6732 | Yes | ||
| 55 | GRB2 | 20149 | 17590 | -3.055 | -0.6496 | Yes | ||
| 56 | SLC9A3R1 | 11250 | 17768 | -3.541 | -0.6131 | Yes | ||
| 57 | PTEN | 5305 | 18200 | -4.996 | -0.5714 | Yes | ||
| 58 | GAB1 | 18828 | 18283 | -5.430 | -0.5052 | Yes | ||
| 59 | STAT1 | 3936 5524 | 18294 | -5.505 | -0.4342 | Yes | ||
| 60 | CSK | 8805 | 18359 | -5.871 | -0.3613 | Yes | ||
| 61 | BLK | 3205 21791 | 18378 | -5.983 | -0.2845 | Yes | ||
| 62 | STAT3 | 5525 9906 | 18455 | -6.760 | -0.2007 | Yes | ||
| 63 | DNM2 | 4635 3103 | 18471 | -6.991 | -0.1106 | Yes | ||
| 64 | MAPK1 | 1642 11167 | 18592 | -9.104 | 0.0013 | Yes |